Molecular dynamics analysis¶
This example shows you how to conduct a 3D molecular dynamics (MD) analysis. MD analysis uses LAMMPS as the default solver. The content will be added in the future.
Creating a MD project¶
Clicking the MD project from the Toolbar or the Menu, A new MD project tree will be presented in the Project Explorer window. Note that MD project is different to the FEM project, and the properties are different as well.
Specifying analysis¶
In this analysis, we set MD Project properties Units to SI, Atom Style to Peridynamics. All the property settings are shown in figure below.
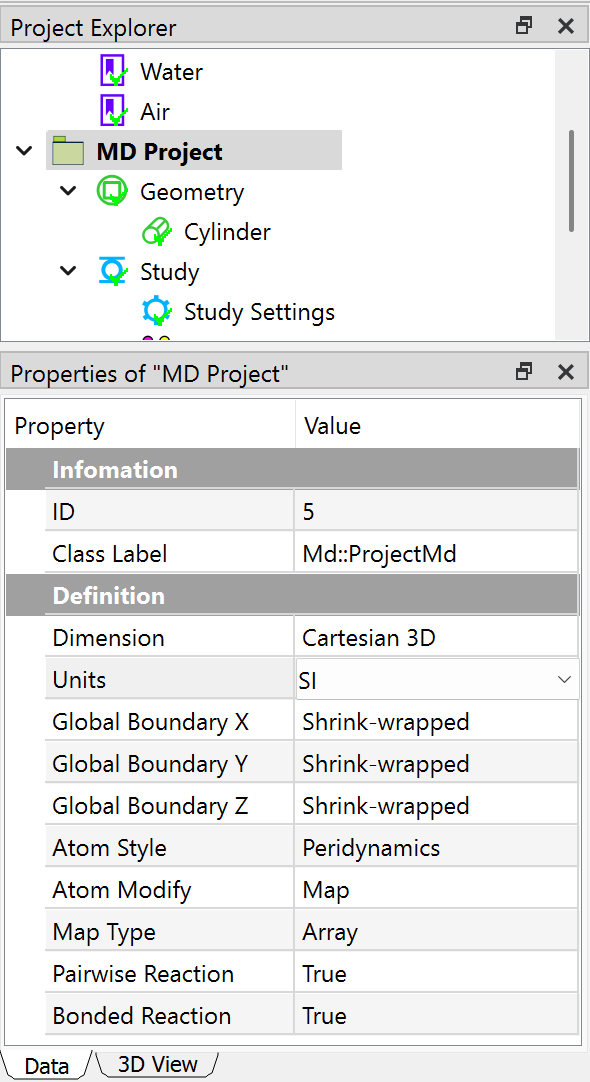
Preparing geometry¶
Next, you can create a cylinder geometry, and define the molecular details in the Property window. As shown in the Figure below, the Lattice Style is set to SC, Lattice Scale is set to 0.0005, Units Value is set to Box. The dimensions for the cylinder is given as well in the Figure below.
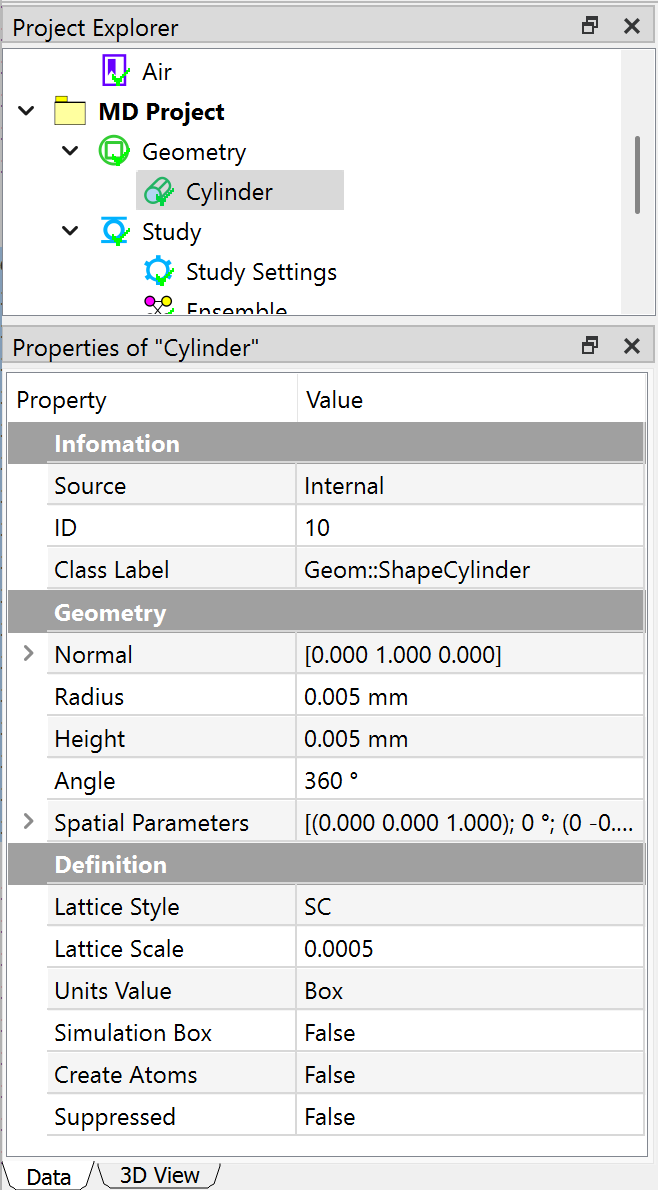
Setting analysis¶
Next, you need to define step time and other properties in the Study Settings object, as shown in the Figure below.
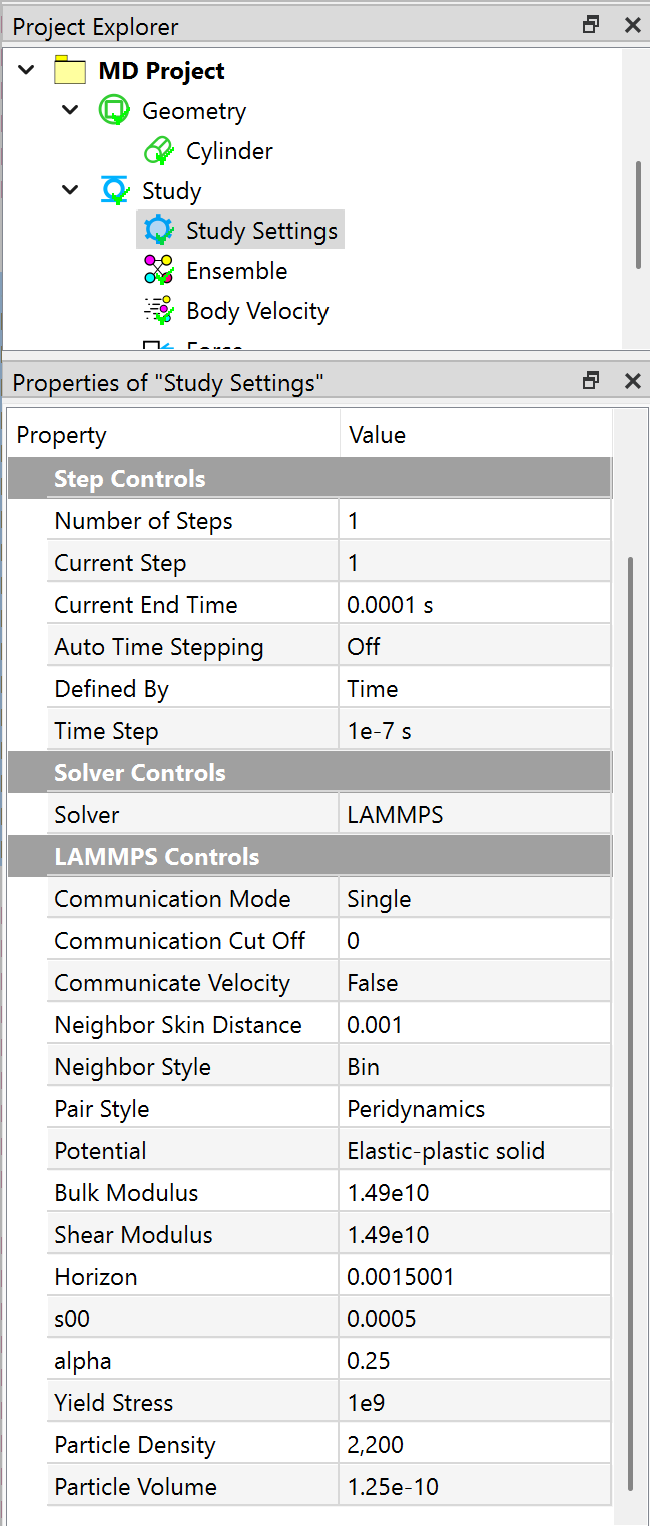
Imposing conditions¶
Next, you impose three conditions, an Ensemble, a Body Velocity, and a Force by clicking the corresponding commands from the Toolbar and MD Menu. In the Properties View of the Ensemble object, choose Microcanonical (NVE) type.
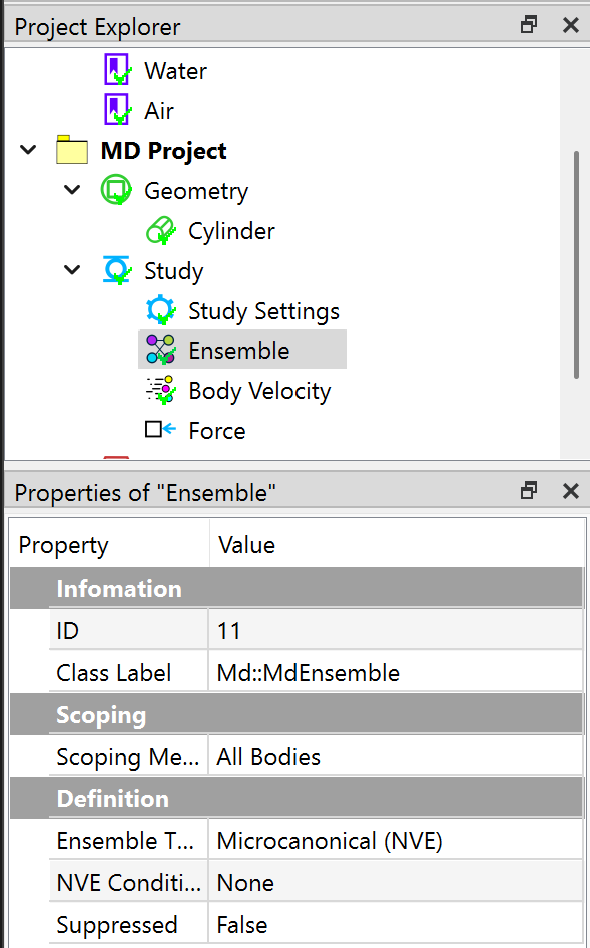
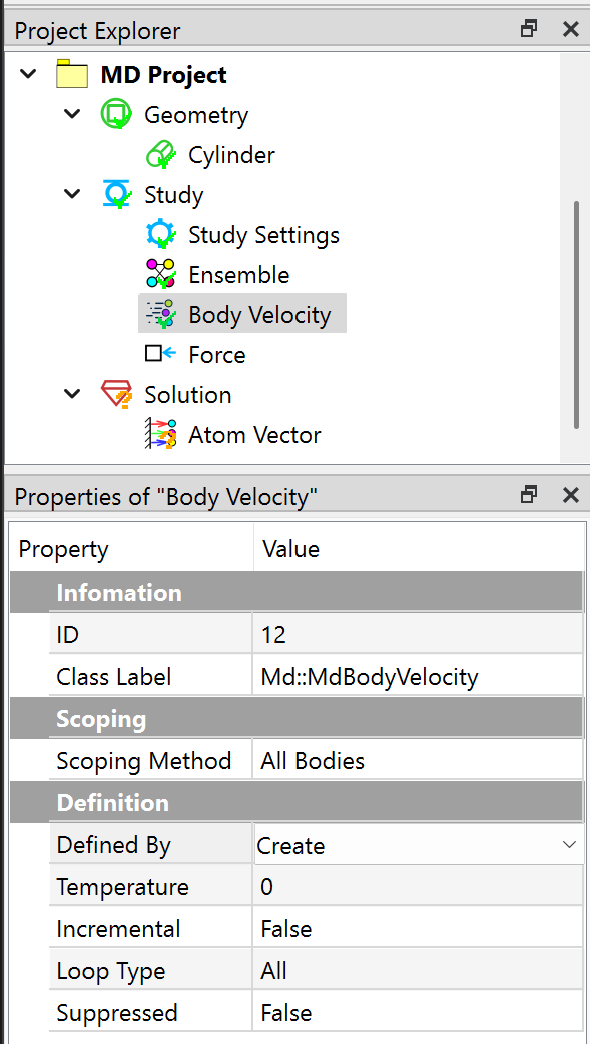
In the Properties View of Body Velocity object, set the Defined By to Create, and Loop Type to All.
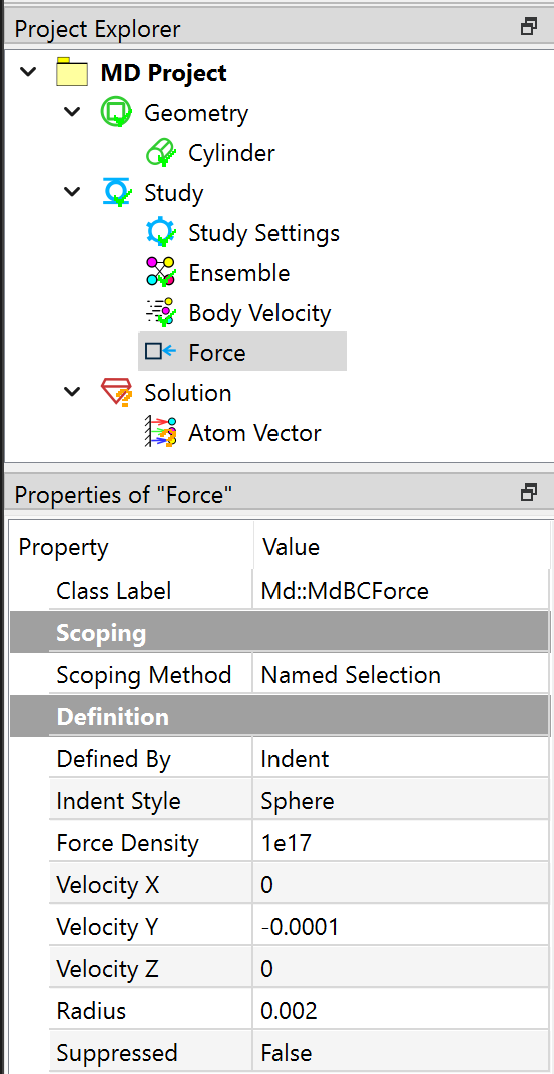
In the Properties View of Force object, set the Defined By to Indent, and Indent Style to Sphere. The other parameters are given in the Figure below.
Solving the model¶
To solve the model, you can click the Compute command from the Toolbar, FEM Menu, or right-click on the Answers object and select Compute command from context menu. Depending on the complexity of the model, the solving process can be completed in seconds to hours. The Output window displays the solver messages and indicates the status of the solving process.